|
|
|
TreeMe
Visualization, Editing and Annotation of Phylogenetic Trees
|
|

|

|
Features:
- Visualisation of large phylogenetic trees (>1000 OTUs)
- Combine trees: unites bootstrap labels of several trees onto a single tree
- Full access to each node: modify node caption, branch caption, branch width, branch length, etc.
- Annotate internal nodes (cluster nodes, clusters)
- Collapse, Expand and Flip clusters
- Re-root trees by using an outgroup
- Define and visualise tags, e.g. arrows, boxes, text labels, etc.
- Automatic replacing of taxon abbreviations (e.g. GenBank accession numbers) by complete names (e.g. by species names) using tab-delimited replacement files (edited with Microsoft EXCEL).
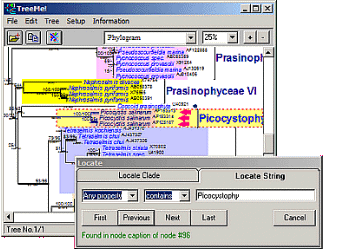
- Fast location of nodes by searching for node names or other properties
- Edit trees: move nodes, delete internal nodes, ...
- Add links to taxa (e.g. to GenBank) and jump to the sequence data with a single mouse click!
- Clipboard functions: Copy tree and insert into other software (Word, Powerpoint, ...)
- Printing of trees
- Imports: NEWICK (Phylip, MrBayes), NEXUS (Paup), ...
- Outputs: NEWICK (Phylip, MrBayes), NEXUS (Paup), Vector Graphics (EMF, Windows Enhanced Metafile)
- ...
Keywords:
tree visualisation, annotation, view, edit
|
|