|
|
|
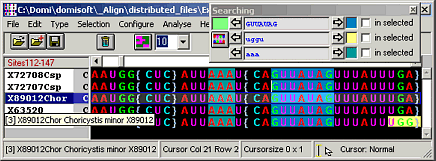
Align
Manual DNA sequence alignment editor
|
|

|

|
Features:
- Alignment of DNA sequence, RNA sequence, protein sequence and other data
- Imports and exports various sequence file formats, e.g. Paup-Nexus, Phylip, Fasta, EMBL, GenBank, DCSE, and others.
- Find/Highlight sequence motives in your molecular sequence alignment (including wildcards ? or *)
- Wizard for automatic processing of sequence alignments for phylogenetic analyses
- Define phylogenetic masks: Which sites are good for phylogenetic analysis?
- Select/Deselect sequences and save selection profiles to define subsets.
- Sort sequences by: accession numbers, taxonomy, arrangement in a phylogentic tree and others
- Several cursor shapes: normal, exclusive (edits all sequences except cursor position), ex- and insertional, left-ended cursor
- Change the cursor to edit several sequences simulataneously
- Analyze sequences and alignments: similarities, p-distances, RASA, ...
- Manipulate aligned sequences: Translate DNA/RNAs to protein using your own codon usage table
- Cut aligned sequences: e.g. remove common positions, where a mask sequence contains special characters
- Create sequences: e.g. consensus sequences, frequency of characters, sites that are only present in selected species (signatures), ...
- Full access to all GenBank fields
- Batch replacement of accession numbers by other fields in foreign files, e.g. replace labels in a tree file. This is not necessary if you have TreeMe.
- ... and many other features.
Keywords:
Manual sequence alignment editor, DNA, RNA, protein, wizard, prepare for phylogenetic analyses
|
|